Makie Helpers
Unit conversions
Many graphical properties are given in pixel or inches. Working with figures for international journals, however, usually requires thinking and specification in metric units. The following function help to convert.
FigureHelpers.cm2inch — Functioncm2inch(x)
cm2inch(x1, x2, x...)Convert length in cm to inch units: 1 inch = 2.54 The single argument returns a value, the multiple argument version a Tuple. Similar to more verbose syntax using Unitful.jl: uconvert(u"inch",x * u"cm").val.
Figures often look well, if the ratio between length and height corresponds to the Golden ratio. So we provide it here.
FigureHelpers.golden_ratio — Constantgolden_ratio ≈ 1.618Two quantities are in the golden ratio if their ratio is the same as the ratio of their sum to the larger of the two quantities.
Makie Layout switching and Figure generation
Generating the same plot for paper and for a presentation, requires adjusting properties such as figure sizes, fontsizes, and graphics format. The following class helps to collect them and provide them to functions below
FigureHelpers.MakieConfig — TypeMakieConfigA collection of figure properties to tailor the same figure either for presentation in a paper or a presentation.
Properties and defaults
pt_per_unit = 0.75px_per_unit = 2.0filetype = "svg"fontsize = 9(point units)size_inches = cm2inch.((17.5,17.5/golden_ratio))(width x height)
The default px_per_unit needs to rescale a png image in ppt to 50%, but this gives better quality.
FigureHelpers.paper_MakieConfig — Functionpaper_MakieConfig(;
target = :paper, filetype = "pdf",
fontsize=9, size_inches = cm2inch.((8.3,8.3/golden_ratio)), ...)
ppt_MakieConfig(;
target = :presentation, filetype = "svg",
fontsize=18, size_inches = (6.65,6.65/golden_ratio), ...)
png_MakieConfig(;target = :png, filetype = "png", ...)Specialized configurations adapted to
paper_MakieConfig: single column Biogeosciences paper (8.3cm)ppt_MakieConfig: half of a wide-screen ppt presentation (13.3 inch), either svg or png Inserting generated svg yields better quality than copy (=png) from VScode preview.png_MakieConfig: same as ppt, but generating png images at 200% scale By default, need to rescale the png to 50% when inserting into presentation.
creating Makie Figures with adjusted sizes and font-sizes
FigureHelpers.figure_conf — Functionfigure_conf(; makie_config)
figure_conf(width2height, xfac=1.0; makie_config)
figure_conf(size_inches = cm2inch.((8.3,8.3/golden_ratio)); fontsize=9, pt_per_unit = 0.75)Creates a figure with specified resolution/size and fontsize for given figure size.
The first two constructors apply the settings from the config, with the possibility to adjust the width/height ration (e.g. to golden_ratio) or multiply the default x-width.
The last constructor sets the properties without referring to a config. It uses by default pt_per_unit=0.75 to conform to png display and save. Remember to divide fontsize and other sizes specified elsewhere by this factor. See also cm2inch and save.
FigureHelpers.figure_conf_axis — Functionfigure_conf_axis(...; makie_config, kwrags...)Call figure_conf but return a tuple (figure, axis). The keyword arguments are passed to the Axis(; kwrawgs...) call.
saving figures to correct format, subdirectory, and dpi-resolution
FigureHelpers.save_with_config — Functionsave_with_config(filename, fig; makie_config = MakieConfig(), args...)Save figure with file updated extension cfg.filetype to subdirectory cfg.filetype of given path of filename. Sets pt_per_unit and px_per_unit according to makie_config.
Shortcuts
FigureHelpers.hidexdecoration! — Functionhidexdecoration!(ax;, label, ticklabels, ticks, grid, minorgrid, minorticks; kwargs...)
hideydecoration!(ax;, label, ticklabels, ticks, grid, minorgrid, minorticks; kwargs...)Versions of hidexdecorations! and hideydecorations! with defaults reversed. This allows to selectively hide single decorations, e.g. only the label.
FigureHelpers.axis_contents — FunctionExtract axis object from given figure position. Works recursively until an Axis-object is returned
Plots of univariate Distributions
using Distributions
d = LogNormal()
fig = density_dist(d; normalize=true, label="normalized");
density_dist!(content(fig[1, 1]), d) # unnormalized into the same figure
fig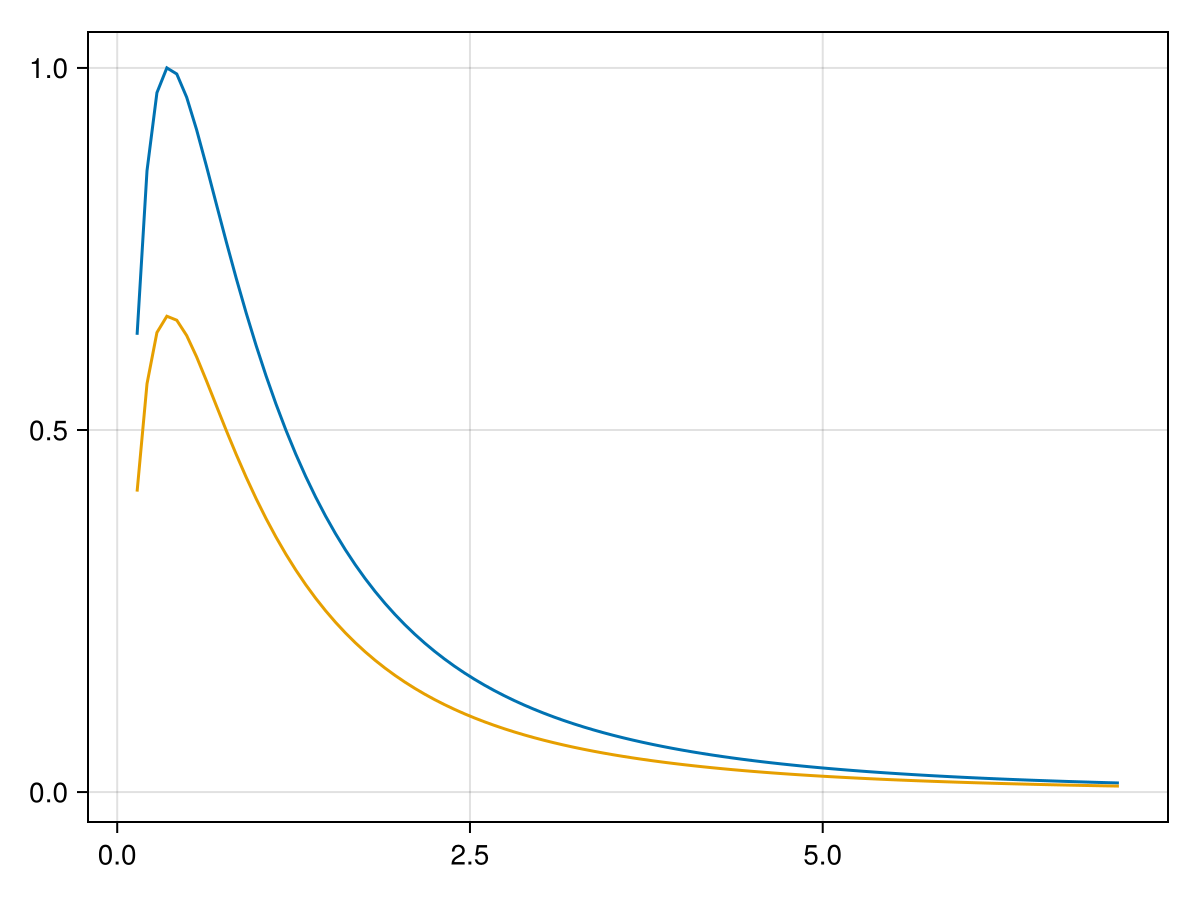
FigureHelpers.density_dist — Functiondensity_dist(d::UnivariateDistribution; ...)
density_dist!(ax, d::UnivariateDistribution; ...)Plot density of univariate distribution, d. The first variant produced a figure. The second plots into given Axis.
Keyword arguments
normalize = false: to scale y-axis to 1 for comparing densities with different spreadprange = (0.025, 0.975): truncate x-axis to focus on main mass of the density
Plots of MCMCChains
Overview: combined trace-plot with density plot.
For linkaxes=true, all the density plots are on the same x-scale.
using MCMCChains
chn = Chains(rand(500, 2, 3), [:a, :b]);
fig = plot_chn(chn; linkaxes=true)
fig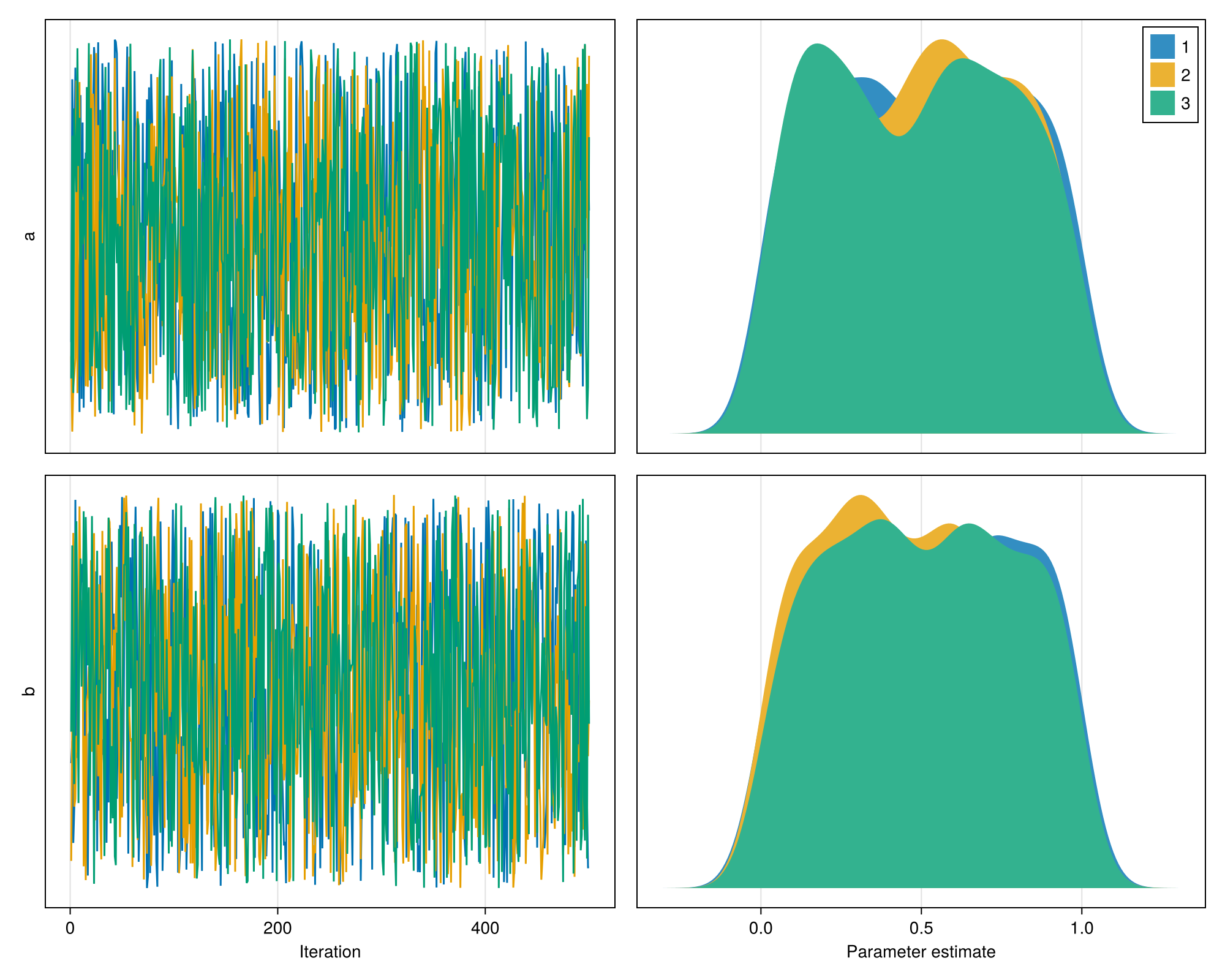
FigureHelpers.plot_chn — Functionplot_chn(fchns::AbstractMCMC.AbstractChains; ... )
plot_chn!(fig::Figure, chns::AbstractMCMC.AbstractChains; ... )Plot MCMCChain lines and density. The first variant produces a figure, the second variant plots into a matrix-grid of axes into given figure.
Keyword arguments
linkaxes=false: link x-axis across density plots of several chainsparam_label="Parameter estimate": x-axis label below lowest density plotparams = names(chns, :parameters): subset of parameters to plot
Tailored density plot
Supports more keyword arguments to adjust color and labels.
using MCMCChains
chn = Chains(rand(500, 2, 3), [:a, :b]);
fig = density_params(chn, ["a","b"], labels=["bla","foo","bar"]);
fig[:, 2] = Legend(fig, content(fig[1,1]), "Chain", framevisible = false)
fig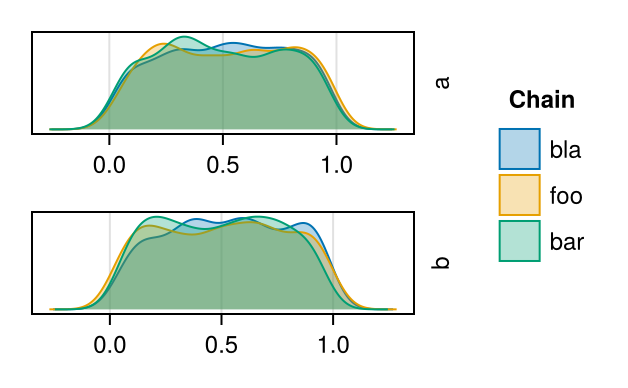
FigureHelpers.density_params — Functionhdensity_params(chns, pars=names(chns, :parameters);
makie_config::MakieConfig=MakieConfig(),
fig = figure_conf(cm2inch.((8.3,8.3/1.618)); makie_config),
column = 1, xlims=nothing,
labels=nothing, colors = nothing, ylabels = nothing, normalize = false,
kwargs_axis = repeat([()],length(pars)),
prange = (0.025, 0.975), # do not extend x-scale to outliers
kwargs...
)
histogram_params(chns, ...)Density/Histogram of several variables of a 3D array, i.e. MCMCChain.
Arguments
column: The axis-grid column into which to plotxlims: indexable of length(pars), providing(lower,upper)bounds tuple for x-limitslabels, colors: names (default chain number) and colors (default palette) of the chainsylabels: column labels (default parameter names)normalize: if true, scale all chain densities to maximum one for each rowkwargs_axis: indexable of keyword arguments to axis for each parameterprange: bounds to the x-axis in percentileskwargs: further keyword arguments tolines!(if normalize) ordensity!
Returns the created figure.
The density plot gives wrong impressions, if probability mass is concentrated at the borders, therefore provide an histogram equivalent.