Hyperparameter tuning with Hyperopt.jl
1. Setup and Data Loading
Load package and synthetic dataset
using EasyHybrid
using CairoMakie
using Hyperoptds = load_timeseries_netcdf("https://github.com/bask0/q10hybrid/raw/master/data/Synthetic4BookChap.nc")
ds = ds[1:20000, :] # Use subset for faster execution
first(ds, 5)| Row | time | sw_pot | dsw_pot | ta | reco | rb |
|---|---|---|---|---|---|---|
| DateTime | Float64? | Float64? | Float64? | Float64? | Float64? | |
| 1 | 2003-01-01T00:15:00 | 109.817 | 115.595 | 2.1 | 0.844741 | 1.42522 |
| 2 | 2003-01-01T00:45:00 | 109.817 | 115.595 | 1.98 | 0.840641 | 1.42522 |
| 3 | 2003-01-01T01:15:00 | 109.817 | 115.595 | 1.89 | 0.837579 | 1.42522 |
| 4 | 2003-01-01T01:45:00 | 109.817 | 115.595 | 2.06 | 0.843372 | 1.42522 |
| 5 | 2003-01-01T02:15:00 | 109.817 | 115.595 | 2.09 | 0.844399 | 1.42522 |
2. Define the Process-based Model
RbQ10 model: Respiration model with Q10 temperature sensitivity
function RbQ10(;ta, Q10, rb, tref = 15.0f0)
reco = rb .* Q10 .^ (0.1f0 .* (ta .- tref))
return (; reco, Q10, rb)
endRbQ10 (generic function with 1 method)3. Configure Model Parameters
Parameter specification: (default, lower_bound, upper_bound)
parameters = (
rb = (3.0f0, 0.0f0, 13.0f0), # Basal respiration [μmol/m²/s]
Q10 = (2.0f0, 1.0f0, 4.0f0), # Temperature sensitivity - describes factor by which respiration is increased for 10 K increase in temperature [-]
)(rb = (3.0f0, 0.0f0, 13.0f0), Q10 = (2.0f0, 1.0f0, 4.0f0))4. Construct the Hybrid Model
Define input variables
forcing = [:ta] # Forcing variables (temperature)
predictors = [:sw_pot, :dsw_pot] # Predictor variables (solar radiation)
target = [:reco] # Target variable (respiration)1-element Vector{Symbol}:
:recoParameter classification as global, neural or fixed (difference between global and neural)
global_param_names = [:Q10] # Global parameters (same for all samples)
neural_param_names = [:rb] # Neural network predicted parameters1-element Vector{Symbol}:
:rbConstruct hybrid model
hybrid_model = constructHybridModel(
predictors, # Input features
forcing, # Forcing variables
target, # Target variables
RbQ10, # Process-based model function
parameters, # Parameter definitions
neural_param_names, # NN-predicted parameters
global_param_names, # Global parameters
hidden_layers = [16, 16], # Neural network architecture
activation = relu, # Activation function
scale_nn_outputs = true, # Scale neural network outputs
input_batchnorm = false # Apply batch normalization to inputs
)Hybrid Model (Single NN)
Neural Network:
Chain(
layer_1 = WrappedFunction(identity),
layer_2 = Dense(2 => 16, relu), # 48 parameters
layer_3 = Dense(16 => 16, relu), # 272 parameters
layer_4 = Dense(16 => 1), # 17 parameters
) # Total: 337 parameters,
# plus 0 states.
Configuration:
predictors = [:sw_pot, :dsw_pot]
forcing = [:ta]
targets = [:reco]
mechanistic_model = RbQ10
neural_param_names = [:rb]
global_param_names = [:Q10]
fixed_param_names = Symbol[]
scale_nn_outputs = true
start_from_default = true
config = (; hidden_layers = [16, 16], activation = relu, scale_nn_outputs = true, input_batchnorm = false, start_from_default = true,)
Parameters:
Hybrid Parameters
┌─────┬─────────┬───────┬───────┐
│ │ default │ lower │ upper │
├─────┼─────────┼───────┼───────┤
│ rb │ 3.0 │ 0.0 │ 13.0 │
│ Q10 │ 2.0 │ 1.0 │ 4.0 │
└─────┴─────────┴───────┴───────┘5. Train the Model
out = train(
hybrid_model,
ds,
();
nepochs = 100, # Number of training epochs
batchsize = 512, # Batch size for training
opt = AdamW(0.001), # Optimizer and learning rate
monitor_names = [:rb, :Q10], # Parameters to monitor during training
yscale = identity, # Scaling for outputs
patience = 30, # Early stopping patience
show_progress=false,
hybrid_name="before"
) train_history: (31,)
mse (reco, sum)
r2 (reco, sum)
val_history: (31,)
mse (reco, sum)
r2 (reco, sum)
ps_history: (31,)
ϕ ()
monitor (train, val)
train_obs_pred: 16000×3 DataFrame
reco, index, reco_pred
val_obs_pred: 4000×3 DataFrame
reco, index, reco_pred
train_diffs:
Q10 (1,)
rb (16000,)
parameters (rb, Q10)
val_diffs:
Q10 (1,)
rb (4000,)
parameters (rb, Q10)
ps: (338,)
st:
st_nn (layer_1, layer_2, layer_3, layer_4)
fixed ()
best_epoch: 0
best_loss: 14.3152846. Check Results
Evolution of train and validation loss
EasyHybrid.plot_loss(out, yscale = identity)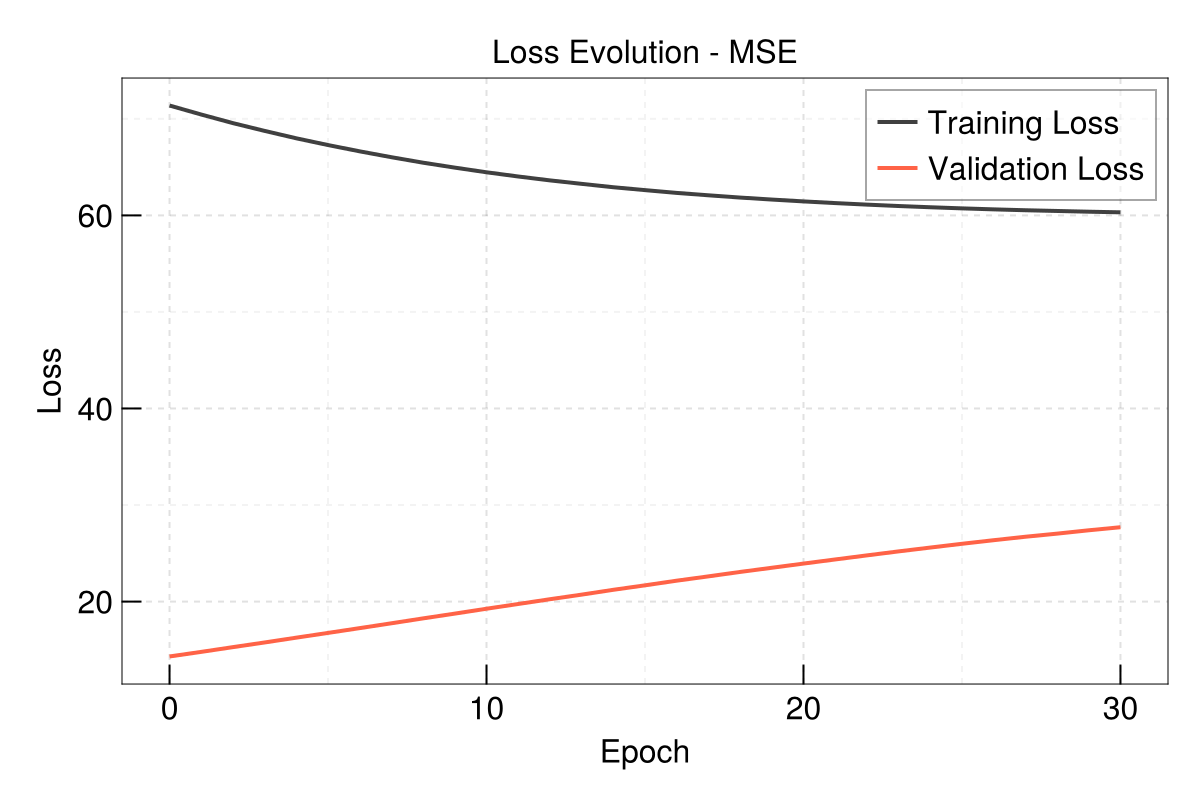
Check results - what do you think - is it the true Q10 used to generate the synthetic dataset?
out.train_diffs.Q101-element Vector{Float32}:
2.0Quick scatterplot - dispatches on the output of train
EasyHybrid.poplot(out)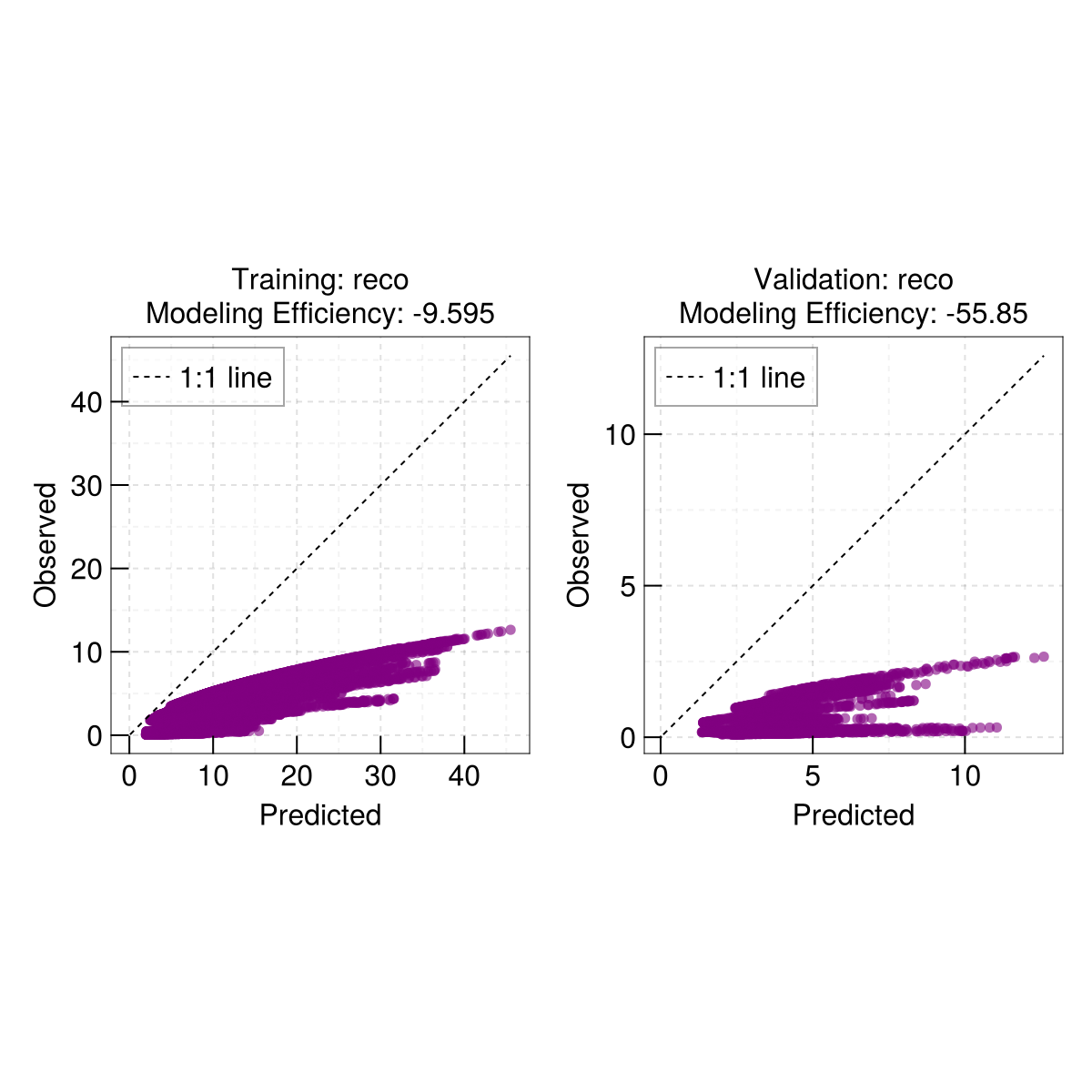
Hyperparameter Tuning
EasyHybrid provides built-in hyperparameter tuning capabilities to optimize your model configuration. This is especially useful for finding the best neural network architecture, optimizer settings, and other hyperparameters.
Basic Hyperparameter Tuning
You can use the tune function to automatically search for optimal hyperparameters. Check Hyperopt.jl for details on algorithms.
# Create empty model specification for tuning
mspempty = ModelSpec()
# Define hyperparameter search space
nhyper = 4
ho = @thyperopt for i=nhyper,
opt = [AdamW(0.01), AdamW(0.1), RMSProp(0.001), RMSProp(0.01)],
input_batchnorm = [true, false]
hyper_parameters = (;opt, input_batchnorm)
println("Hyperparameter run: ", i, " of ", nhyper, " with hyperparameters: ", hyper_parameters)
# Run tuning with current hyperparameters
out = EasyHybrid.tune(
hybrid_model,
ds,
mspempty;
hyper_parameters...,
nepochs = 10,
plotting = false,
show_progress = false,
file_name = "test$i.jld2"
)
out.best_loss
end
# Get the best hyperparameters
ho.minimizer
printmin(ho)
# Train the model with the best hyperparameters
best_hyperp = best_hyperparams(ho)(opt = AdamW(eta=0.01, beta=(0.9, 0.999), lambda=0.0, epsilon=1.0e-8, couple=true), input_batchnorm = true)Train model with the best hyperparameters
# Run tuning with specific hyperparameters
out_tuned = EasyHybrid.tune(
hybrid_model,
ds,
mspempty;
best_hyperp...,
nepochs = 100,
monitor_names = [:rb, :Q10],
hybrid_name="after"
)
# Check the tuned model performance
out_tuned.best_loss0.0011677144f0Key Hyperparameters to Tune
When tuning your hybrid model, consider these important hyperparameters:
Optimizer and Learning Rate: Try different optimizers (AdamW, RMSProp, Adam) with various learning rates
Neural Network Architecture: Experiment with different
hidden_layersconfigurationsActivation Functions: Test different activation functions (relu, sigmoid, tanh)
Batch Normalization: Enable/disable
input_batchnormand other normalization optionsBatch Size: Adjust
batchsizefor optimal training performance
Tips for Hyperparameter Tuning
Start with a small search space to get a baseline understanding
Monitor for overfitting by tracking validation loss
Consider computational cost - more hyperparameters and epochs increase training time
More Examples
Check out the overview page for additional examples and use cases. Each project demonstrates different aspects of hybrid modeling with EasyHybrid.
 Bernhard Ahrens
Bernhard Ahrens